Abstrict
This review article is aimed at providing an outline of the present information base available on the applications of CRISPR/Cas9 in cancer therapy and the challenges raised by its implementation. We will first explain the mechanism of action and other key components pertaining to the functioning of CRISPR/Cas9 technology. Further, we will provide case studies that have had some success in pre-clinical and clinical applications of CRISPR/Cas9. However, there are several challenges that prevent the wide application of this very promising approach of CRISPR/Cas9 in cancer therapy. Among them are off-target effects and unintended mutations, difficulties in delivery, targeting specificity, etc. Specificity enhancement, improvement in delivery systems, and solutions against regulatory challenges have been described.
Keywords
Recombinant DNA Technology, CRISPER/Cas-9, Applications, Challenges, Genome, Genome Engineering
Introduction
In the 1970s, recombinant DNA technology was discovered; this represented a key milestone in biological research as it provided molecular biologists with all new opportunities. Such technology in fact made it possible to manipulate DNA molecules and study genes, as well as develop aspects of modern medicine and biotechnology. The modern era of innovative research in the field of biology begins with discovery and development, respectively, which we have already available through recently developed genome-engineering techniques. Now instead of studying DNA in isolation from the genome, scientists are able to directly manipulate or even modulate a specific genetic sequence finally, anywhere in their native milieu among many different taxa (Patrick D. Hsu, 2014). This provides a unique opportunity for researchers to query the system-level functional organization of this genome and discern causal genetic variations (Patrick D. Hsu, 2014).
Genome engineering is generally used to describe any technology that directly alters the genetic material of a cell, usually as part of an effort to bio streamline/enhance its known functions. The simplicity and efficiency of performing these modifications, especially in mammalian and eukaryotic cells have implications for basic science, biotechnology as well as medicine. (Patrick D. Hsu, 2014).
In the context of life sciences research, technologies that can remove a section of DNA and fill in new sequences into cells or organisms are important tools for understanding individual genes and regulatory elements. Multiplexed editing also greatly extends this capability and can be used to interrogate a wider range of genomic networks, or protein neighbourhoods than previously possible. In the same way that transcriptional regulation or control over chromatin states in a targeted fashion helps us understand how genetic material is packaged and employed within cells, changes to genome architecture are important because we can infer on an organism level about function. (Patrick D. Hsu, 2014).
Through the exact modification of genetic components and behavioral elements, biotechnology enables the development or reverse engineering of these practical biological systems. These methods might involve creating crops that are resistant to infection or optimizing the processes via which industrially relevant organisms produce biofuel. Additionally, advocated genome engineering for drug discovery and medicinal therapeutics. The cumulative effects that affect complicated polygenic illnesses can be modeled and new therapeutic targets can be found by simultaneously perturbing numerous genes. Furthermore, harmful mutations may one day be repaired using genome editing in order to facilitate human gene therapy (Patrick D. Hsu, 2014).
A variety of genome-modifying approaches based on nucleases with programming capabilities have surfaced recently to solve these issues, enabling the efficient and focused modification of different eukaryotic species, especially mammals. Among the current generation of genome editing tools, the class of RNA-guided endonucleases called Cas9 is one of the fastest developing. It is derived from the microbial adaptive immunity system CRISPR (clustered regularly interspaced short palindromic repeats). With the use of a brief RNA guide, Cas9 may be easily guided to nearly any chromosomal site (Patrick D. Hsu, 2014). However, does Genetic Engineering technology contribute to Cancer therapy?
100 years before, cancer was not very common, but in the last few years, its influence in the disease world has increased a lot due to more chances of dietary and lifestyle changes and most importantly expectancy of life. It has become one of the scary diseases of the last century and its occurrence is increasing still. The consequences are quite detrimental that every single out of dur individuals faces this danger. Especially in India, more than 1.1 million cases are reported annually. On the other hand, the number exceeds 14 million. People are continuously susceptible to different cancer-inducing agents which are also called carcinogens (Roy & Saikia, 2016).
Cancer is the irregular division of cells which is detrimental to health. They can rise from any part of the body and they are made from small cells and have lost the power to inhibit division due to which division continues and ultimately becomes tumor. Sometimes cancer may be detected accidentally during the conduct of lab tests and for unknown reasons. Usually, cancer cells need to reach a level of 1 cm, which may composed of 1 million cells, before it can be found as a disease. This stage normally displays or manifests as a tumor, lump mass, etc. Some cancer types like lymphomas do not produce mass but can be find by lab tests (Roy & Saikia, 2016).
It is possible that the body's immune system fails to recognize and destroy these newly generated degenerative cells when they are still in trace amounts, rather than that a healthy cell turning malignant is the main cause of cancer. People with weakened immune systems are more likely to get cancer because of things like aging, long-term stress, chronic illnesses, prior chemotherapy, and abusing medications like analgesics, corticosteroids, and antibiotics. Cancer's impact on the global scene has resulted in notable rates of morbidity and mortality. However, advancements in genetic engineering have led to a significant improvement in cancer therapy over time (Roy & Saikia, 2016).
For example, immunotherapy, RNA modifications, RNA-directed degradation techniques, and others have gradually reduced the impact of cancer. Similarly, CRISPR/Cas-9 shown in figure-1, one of the most remarkable genetic engineering technologies, is used as a therapeutic tool to suppress or degrade mutated cancerous cell DNA through complex pathways (Roy & Saikia, 2016).
Since the invention of the DNA double helix, scientists and medical experts have contemplated the possibility of modifying the genomes of cells and organisms at specific locations. Early genome editing methods were based on the notion that DNA sequences might be identified by their unique locations. Studies on natural DNA repair pathways and DNA recombination mechanisms in bacteria and yeast have demonstrated that cells possess endogenous machinery capable of repairing double-strand DNA breaks (DSBs) that would otherwise be lethal. Consequently, methods for accurately cleaving DNA at specific sites were recognized as effective strategies for targeted genomic editing. (Doudna, 2014).
It is crucial to investigate new paths to progress cancer medicines in light of the aforementioned difficulties as well as the need for tailored therapies, higher survival rates, and better treatment tolerance. Numerous cutting-edge systems, like the CRISPR/Cas system, have surfaced in this field. Compared to traditional genome modification methods, this adaptable technology can be applied as a tool for altering genes to precisely alter DNA sequences at particular genomic locations. The goal of current work is to find clinical uses for this easily accessible preclinical instrument, which has produced encouraging findings. Consequently, the benefits and difficulties of CRISPR/Cas-9 in cancer therapy are covered in this review study. (Dimitrios Stefanoudakis, 2023).
Figure 1
CRISPER/Cas-9

Applications of CRISPR/Cas9 in Cancer Therapy:
Epigenetic Basis of Cancer and Challenges of Conventional Therapies
Cancer is a result of many different types of genetic and epigenetic changes in the cell that finally lead to uncontrolled cell growth and tumor formation. Epigenetic alterations at the levels of DNA methylation, histone modifications, and noncoding RNA regulation are key events in the emergence and spread of malignancies.
Perhaps among the most important defects of most conventional cancer therapies, including chemotherapy, is their relative non-selectivity, which does not only kill cancerous cells but also normal cells. Further, continuous administration would result in resistance to such therapies developed by cancer cells themselves. Therefore, more specific and precisely targeted therapeutic approaches are urgently required in cancer treatment.
Identification of Novel Targets
The investigators are searching for new molecular targets to overcome these limitations of conventional therapies. Targeting tumor suppressor genes and mutant oncogenes in cancer cells is one such strategy. When these genes are unregulated, it can lead to the unchecked proliferation and survival of cancer cells.
CRISPR-Cas9-Mediated Regulation of Tumor Suppressor Genes
The new genome-editing tool, known as CRISPR-Cas9, is such a way to aim and alter the expression of those genes implicated in cancer. It has been shown that expression of tumor suppressor genes, such as hBax, E-cadherin, and p21, can be efficiently controlled by CRISPR-Cas9 to finally suppress the proliferation of cancer cells and induce apoptosis.
Overcoming Chemotherapeutic Drug Resistance
This is a major challenge in the treatment of cancer, often mediated by mechanisms such as overexpression of drug efflux pumps. Knockdown by CRISPR-Cas9 at genes involved in drug resistance—including MDR1—reconstitutes cellular sensitivity to chemotherapy drugs and hence is a potential way to circumvent this problem.
Targeting Other Cancer-Associated Genes
Other genes involved in cancer, such as SHCBP1 and KLHDC4, are amenable to the genome editing tool CRISPR-Cas9. Knockout of genes by CRISPR-Cas9 has reportedly shown much promise in inhibiting the growth of cancerous cells and inducing apoptosis
CRISPR-Cas9 for Cancer Model Development
The introduction of CRISPR-Cas9 technology greatly improved the creation of cancer models created by genetic engineering in mice with the capability to replicate the state of disease in humans. This way, the models can be harnessed for the study of cancer progression from metastasis and response to therapy in a controlled environment, thereby hastening the discovery of new treatments.
Gene Editing-Based Immunotherapy in Cancer
CRISPR-Cas9 gene-editing technology has revived the innovative approach for immunotherapy, inside which lie the chimera antigen receptor (CAR) T cell therapies and genetically modified T cell receptor (TCR) therapies against cancer. Thus, gene modification by the new gene-editing CRISPR-Cas9 approach has meant the killing of specific targeted antigens on carcinoma cells, hence their death. Similarly, having fewer side effects, using CRISPR-Cas9 in TCR therapy shows new hope for cancer.
Future Directions and Clinical Trials
Clinical trials are underway to investigate the use of the targeted nucleases, namely CRISPR-Cas9-based therapies in various cancers. Some of these trials are trying to establish the safety and efficiency of CRISPR-Cas9-mediated interventions that would argue for their full-scale adoption in clinics.
Figure 2
PD-1 Receptor Gene Editing
It has been interrogated that engagement of receptors, including programmed death-1, modulates immune responses. It meant that PD-1 receptor gene disruption through the CRISPR-Cas9 system in T cells was followed by augmented immune responses against cancer cells.
Review of CRISPR/CAS9 in Tumor Diseases
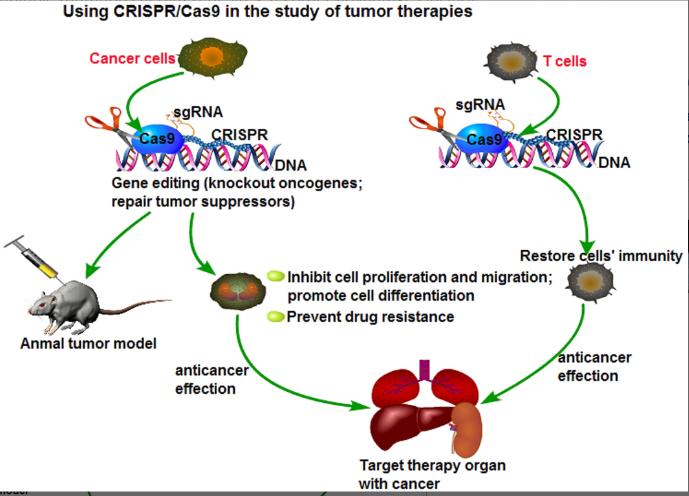
Applications of CRISPR/Cas9 in tumor therapies involve the use of accurate gene editing to treat diseases. This method of gene editing can make permanent changes by removing unwanted genes or adding protective genes. It allows efficient and easy production of homozygous mutants in a much better way than the previously known methods like ZFN and TALEN, and also the introduction of multiple mutations at different sites. What is more, CRISPR/Cas9 is of low cytotoxicity. Such an advantage makes it prevalent and now has many applications. The application is mostly concentrated on genetic diseases, viral infections, and tumors. In the research of tumors, CRISPR/Cas9 is also frequently employed to investigate therapeutic interventions and the mechanisms underlying tumor development. Interest areas include genes connected to cancers, the creation of animal models for tumors, medication resistance genes, phenotypic correlations, and cancer gene therapy.
Target editing of microRNA (miRNA)
Modifying specific microRNAs, or miRNAs, is a field of research that is gaining popularity. Small non-coding RNA molecules (miRNAs) attach to the 3?-untranslated region of target genes to regulate gene expression at both the transcriptional and post-transcriptional stages. Based on their ability to modulate target gene expression, miRNAs have been shown to have a close relationship with tumor formation. They regulate important mechanisms such as the cell cycle, metastasis, apoptosis, and treatment resistance. Enhancing the effectiveness of tumor treatment can be achieved by targeting and altering certain miRNAs within tumor cells, especially those that advance the tumor, and then suppressing them to prevent tumor development and overcome chemotherapy resistance. A series of studies including the knockout of miR-17 in the colorectal cancer cell line HT-29 and subcutaneous injection into nude mice showed that CRISPR/Cas9 may create a persistent phenotype by targeted miRNA deletion. The transplanted tumor tissue showed a stable gene-phenotype after two weeks. Large drops in the levels of miR-17, miR-200c, and miR-141 after intervention further demonstrated that CRISPR/Cas9 is effective at targeting and downregulating specific miRNAs in human colorectal cancer HCT116 cells. (Rasul, 03 March 2022).
Anticancer application of CRISPR/Cas9 gene editing
During the onset and development of cancer, a wide range of genes, including oncogenes, tumor suppressor genes, genes linked to stem cells, chemo-resistant genes, and metabolic genes, are altered or dysregulated. The main strategies used in cancer treatment to stop cancer cells from growing and spreading are mutation correction and the restoration of normal gene expression patterns. The CRISPR/Cas 9 gene-editing method has been widely used in cancer research since its invention, with promising results. Recently, Georgiadis et al. demonstrated how to create T cells immune to fratricide by substituting lentiviral-mediated CARs that target CD3 or CD7 for the TCR/CD3 and CD7 (Wang H, 2016 June). Table 1 presents a noteworthy compilation of target genes, tumor types, and data illustrating the degree to which CRISPR/Cas9 corrects these genetic alterations. Targeting cancer-causing genes with the CRISPR/Cas9 system could be a practical application given the encouraging outcomes of preclinical research (Zhang JH, 2016 Apr;7)
Figure 3
Challenges in IN VIVO Technologies for gene editing of CRISPR/CAS9
Safe and effective ways to deliver quality-altering chemicals to target growth cells and their metastases are needed in order to assess the usage of quality-altering tools like the CRISPR/Cas9 framework in the future. Beneficial ex vivo quality alteration has mostly been performed on hematopoietic progenitors or lymphocytes thus far. More uses of CRISPR-based therapy will only be possible with the development of efficient in vivo delivery techniques in big cells. New, widely used, non-viral frameworks could be developed or implemented to get over these extra transportation constraints. Lentivrons, adenoviruses, and adeno-related infections (AAVs) are examples of viral delivery mechanisms for CRISPR/Cas9 components. (Marta Martinez-Lage, 2018)
As of right now, AAVs are the most advanced technique available for in vivo quality delivery. AAV is without a doubt an excellent vehicle for top-notch care for the following reasons: I AAV is not known to cause any human illnesses; (ii) there is a large variety of known serotypes for diseases that affect various cell types; and (iii) AAV nearly never results in a safe reaction. AAVs have also been successfully utilized in animal models, and late-stage clinical trials have tested the viability and well-being of these vectors. AAVs are currently the best technology available for in vivo quality delivery. AAV is unquestionably a fantastic vehicle for high-quality care for the following reasons: I There are no known illnesses associated with AAV in humans; (ii) a wide range of known serotypes for diseases affecting different cell types; and (iii) AAV almost never causes a safe reaction. Additionally, While AAVs have proven effective in mouse models, one of their main disadvantages is their small bundling size. This means that multiple infections are required to transfer all of the CRISPR/Cas9 components (Cas9, sgRNAs, and potentially significant contributing DNA), which reduces the alteration's efficiency. (Marta Martinez-Lage, 2018).
In contrast to several other techniques, using AAV allows for a persistent articulation of CRISPR regions in modified cells, potentially increasing the risk of resistant reactions or unfavorable off-target genomic effects. Both lentivirus and adenovirus have the ability to contaminate both dividing and non-dividing cells, similar to AAV. Adenoviruses, on the other hand, do not integrate into the recipient cell's DNA, in contrast to lentiviruses and AAV at low recurrence. Lentivoviral tropism can also be changed by other viral proteins, such as the G-protein of vesicular stomatitis infection (VSVG). However, there are certain disadvantages to using lentivirus and adenovirus because they cause powerful immune reactions (Marta Martinez-Lage, 2018)
As mentioned, these viruses are very adaptable particles since they may be used in vitro, ex vivo, or in vivo, which simplifies security and viability testing. An alternative approach to address these problems instead of depending solely on viral delivery would be the use of short, preassembled Cas9 RNP structures or non-viral vector delivery. Among the non-viral strategies are lipid nanoparticles/liposomes, gold nanoparticles, and inorganic nanoparticles [81]. Lipid nanoparticles have been employed for some time now as delivery systems for a variety of other particles in cells. They may allay security and immunogenicity worries because they don't include any well-known components (Marta Martinez-Lage, 2018).
Additionally, they can be used ex vivo, in vivo, and in vitro. An additional advantage of delivering CRISPR components via nanoparticles is their high stacking limit, which eliminates the consequences of cautious CRISPR/Cas9 articulation and the possibility of genomic coordination. In U2OS human osteosarcoma cell xenografts, the transport of Cas9-sgRNA RNP assemblies by nanoparticles has been documented [92]. Furthermore, it has been shown that a distinct delivery method using gold nanoparticles complexed with cationic endosomal problematic polymers and bound to DNA can transfer donor DNA and Cas9 RNP structures. This may promote HDR in order to treat the DNA changes linked to Duchenne's severe dystrophy in mice. Safe carriers for pharmacological and superior delivery applications are offered by gold nanoparticles. The group is stabilized by the gold core, and surface properties such as hydrophobicity and charge can be modified by the monolayer. Although further testing is necessary, this approach offers a potential delivery system for CRISPR components. Since inorganic nanoparticles, such as carbon nanotubes and exposed mesoporous or thick silica nanoparticles, have been used for comparable applications in the past, they are unquestionably frequent candidates for CRISPR component transporters. Furthermore, inorganic nanoparticles are simpler to make and have consistent long-term piece, size, and reliability. (Marta Martinez-Lage, 2018)
Delivery Challenges
Although CRISPR/Cas9 has been widely utilized to modify gene quality, there are still a number of problems with it because of side effects, difficulty in application, and security concerns. Regarding CRISPR-based quality treatment, the challenges associated with in vivo transfection methods are emphasized. High transfection productivity, a unique focus on the limit, and ease of large-scale manufacturing are essential components of a complete CRISPR/cas9 transfection process. However, these goals are still far from being accomplished by the existing techniques.58 Although real CRISPR/Cas9 transfer methods are highly ineffective when applied in vivo, they are mainly employed in vitro. Numerous methods have been widely used, including electroporation, microinjection, ultrasonic-driven nanomotors, nanofluidic/nanofluidic systems, and nanocarriers. Electroporation remains the primary choice in several investigations for the application of CRISPR/Cas9 in human undifferentiated cell quality alteration. CRISPR/Cas9 can, however, be transferred in vivo using certain legitimate methods. One novel technique of delivering CRISPR, hydrodynamic infusion (HDI), was mentioned; however, due to the risk of damage during transfection, its application is restricted. Researchers are administering CRISPR/Cas9 via a range of transporters, in addition to the techniques themselves. (Mohadeseh Khoshandam, 2024)
Two steps make up the CRISPR/Cas9 transfection process which categorizes in addition to the types of transporters: viral transfection strategies and non-viral approaches. Because of the increased transfection rate, viral vectors are more prevalent for CRISPR/Cas9 transfection. Because of its broad serotype, low immunogenicity, and toxicity, adeno-related infection (AAV) is the most well-known vector for effective treatment both in vivo and in vitro. Nevertheless, its small conveyance limit (about 4.5–5 kb) should be moved along. Lentivaunts (LV) and adenoviruses (AdV) move the extra genetic components and have a better, higher, and more grounded limit than AAV. Because of this, one of AdV's main advantages over LV and AAV is its ability to transfect a larger range of cells. Sometimes, larger LV and AdV dimensions can cause humoral reactions and definitely cell immunity. More virus vectors with distinct traits have just now surfaced. (Mohadeseh Khoshandam, 2024)
For instance, baculovirus vectors have a higher capacity for transfection, Sendai infection vectors are more skilled at infecting a larger range of cells, and EBV vectors are more able to stay stable in cells. Even if these vectors are now employed in a variety of contexts, they can still be utilized in vivo with additional simplicity. Regarding non-viral strategies, liposomes are sometimes used due to their widespread advertising. A few researchers attempted to transfer CRISPR/Cas9 in different models using gold nanoparticles (AuNPs). Several non-viral vectors, including Lipofectamine RNAiMAX, 136 PolyJetTM In Vitro DNA Transfection Reagent, and X-tremeGENE HP DNA Transfection Reagent, are appropriate for in vitro or ex vivo testing. For in vivo transfection, a lot of researchers are concentrating on finding and combining non-viral vectors with high productivity and minimal cytotoxicity. (Mohadeseh Khoshandam, 2024)
These strategies include regular nano-sized clusters (such as self-gathered micelles and polyethylene glycol-changed cationic lipid nanoparticles for CRISPR/Cas9 plasmid conveyance) and DNA nanostructures and phosphorus nanosheets. Methods for CRISPR/Cas9 transfection include KASIM transfection (e.g., a K9 protein crossbreed), cell-infiltrating peptides (CPPs)-intervened transfection, and receptor-interceded transfection (e.g., plasmids of CRISPR/Cas9 recognized by the folate receptor). Wang synthesized PEGylated nanoparticles based on cationic ?-helical polypeptides to deliver sgRNA and CRISPR/Cas9 plasmids. Furthermore, a few delivery methods were developed, such as the peptide-changed cationic liposome that R8-dGR used to transport sgRNA and CRISPR/Cas9 plasmids, as well as the near-infrared change-initiated structure. The fastest rapid rate of Cas9/sgRNA transfection using a four-part formula [DOTAP (1,2-dioleoyl-3 trimethylammoniumpropane)/DOPE/cholesterol/Chol-Stake (cholesterol-polyethylene glycol)] resulted in a 39% quality changing productivity to delete GFP reporter, according to a recent publication. Nevertheless, there are a few issues with non-viral approaches, such as endosomal escape and move barriers. (Mohadeseh Khoshandam, 2024)
Unlike other vectors, the CRISPR/Cas9 movement also heavily depends on the statuses of the freights. Cas9 is inserted into the DNA or mRNA state in conjunction with sgRNA and the design arrangement. Yin and colleagues transported Cas9 mRNA by lipid nanoparticles due to quality-altering efficiency, while moving sgRNA and creating the design separately using AAV. Furthermore, using hybridization techniques, It is also possible to successfully transfect cells with Cas9 proteins. This minimizes the possibility of genomic respectability and decreases the influence of off-target effects because of the short half-life of the Cas9 protein, making it a safer approach to high-quality therapy. Therefore, fewer safe reactions will occur the lower the transfection vectors' conveying limit is; nevertheless, a higher conveying limit is necessary to transfer more CRISPR. However, because of their size flexibility, biocompatibility, and biodegradability, multifunctional fantasy peptides have shown potential for quality delivery and may be interesting for CRISPR-based quality change. (Mohadeseh Khoshandam, 2024)
Off-Target Effect of CRISPR/CAS9
Using a designed nuclease results in a form of unintentional and ambiguous genetic change known as "off-target genome altering." For DNA groups with even three to five base pair jumbles in the PAM-distal region of the sgRNA-directing succession, potential off-target cleavage action might still occur, even though the recurrence, kind, and extent of this hereditary change were not made clear. (Asmamaw Mengstie, 2023)
Noncanonical PAMs and a range of unique nucleotides from on-track destinations are present in off-target regions; gRNAs can withstand bungles far more easily at their 5? ends than at their 3? ends. (Asmamaw Mengstie, 2023)
In the improbable case that at least one of them exists, Cas9 activation may be prevented by confusion in the seed region. If there are at least three confounds, DNA groups attach to Cas9, preventing cleavage and discouraging HNH adaptation. The off-target activities of gRNAs are usually influenced by their patterns and configurations within the system.2. Off-target genome editing causes confusion and decreases CRISPR-Cas9 usage, and it may also raise questions about the validity of scientific findings involving gene activities. Because of this, the effect may result in harmful events like cytotoxicity, safe reactions, and unwanted DNA damage (Asmamaw Mengstie, 2023).
When Cas9 is bound to a specific sgRNA and edited at a location other than its target sequence, this is known as off-target editing. Unexpectedly significant effects could include immunological response, oncogene activation, mutation, and the activation or silencing of undesirable genes (Asmamaw Mengstie, 2023).
PAM Limitation
The PAM succession, as it was examined, is essential to CRISPR/Cas9 and heavily dependent on Cas9 explicitness. PAM succession reduces the flexibility of CRISPR/Cas9 with various frameworks and limits the design of sgRNA. new PAM groupings have been discovered in spite of the fact that new CRISPR types are being discovered. The required PAM expansion really affects the sgRNA design under a few different circumstances. Therefore, developing and increasing the use of CRISPR/Cas9 requires a designable PAM. (Khoshandam, Mohadeseh, 2024)
CRISPR-Cas systems would target their CRISPR arrays in the absence of the PAM requirement, which could trigger a disastrous autoimmune reaction. A PAM is necessary for almost all CRISPR nucleases, in one way or another. The identified PAM sequences, however, varied greatly from one another and were not shared by all Cas nucleases. They have distinct lengths, complexity, orientations, and distances from the target. (Mohadeseh Khoshandam, 2024)
Because of this restriction on our capacity to use CRISPR to target any sequence, there have been numerous attempts to loosen the PAM requirement, even to the point where almost any sequence would be considered a PAM. Methods for determining PAM have also been essential in clarifying the sequences that each nuclease recognizes. (Mohadeseh Khoshandam, 2024)
Various PAM sequences are recognized by distinct Cas enzymes, including variations of the standard Streptococcus pyogenes Cas9 (SpCas9). Even yet, a sizable portion of the genome cannot be edited and is more likely to result in off-target changes. Applications requiring high-resolution genome targeting have a major constraint in the form of the PAM requirement. (Mohadeseh Khoshandam, 2024)
Immune Response
Although there haven't been many investigations on the extremely resistant reactions that Cas9 causes, antibodies against the protein have been found in human bodies on a large scale152. This emphasizes the possible danger of interference with CRISPR/Cas9-based high-quality therapies. (Chamberlain, 2018)
Researchers focus more on the immunogenicity of prominent vectors because the human body may have previously been exposed to them. All things considered, the safe reactions arising from the CRISPR/Cas9 quality-altering structure represent a significant risk factor in the development of CRISPR-based quality treatment in vivo. (Chamberlain, 2018)
Anti-Cas9 antibodies show that intracellular bacterial proteins have been exposed to an invulnerable framework during disease, however, they might not be useful against treatment-resistant reactions. Antibodies play a crucial role in engulfing viruses and tiny organisms to prevent them from penetrating cells and designating them for the immune system to destroy. (Chamberlain, 2018)
They can also stamp host cells that are malignant or contaminated and express the target protein on their surface. However, in general, an intracellular protein-specific antibody will not directly result in the death of a cell that is producing that protein. Rather than antibodies, cell-resistant responses—more precisely, CD8+ cytotoxic T lymphocytes (CTLs)—intervene to halt the killing process (Chamberlain, 2018).
Therefore, the rate at which cells become immune to Cas9 is what needs to be considered the most. The presence of anti-Cas9 immune system microorganisms in these donors5 shows two things: first, that these white blood cells are successfully introducing Cas9 through major histocompatibility complex (MHC) particles; and second, those lymphocytes are capable of responding to Cas9.. (Chamberlain, 2018)
When these lymphocytes are activated in conjunction with proinflammatory "risk" signals during a bacterial infection, contaminated cells can be destroyed by CTLs. The Cas9-receptive CD8+ immune system bacteria identified by Charlesworth et al.5 release interferon-?, suggesting they could eliminate Cas9-communicating cells after receiving high-quality therapy, even though they haven't been directly tested as a lethal measure. Overall, the safe framework may destroy the very cells that CRISPR-Cas9 corrected, making the therapy ineffective. (Chamberlain, 2018).
Methods of Overcoming Cancer by Crisper/Cas9
Although cancer is still the world's greatest cause of death, advances in diagnosis, prevention, and treatment provide hope. Despite its effectiveness, radiation and chemotherapy can have serious adverse effects. Comprehending the biology of cancer is essential to creating more effective medicines. The study of the cancer genome can now be done more thoroughly and economically because of recent developments in sequencing technology. Treatment choices can be more individually tailored for each patient when genetic and transcriptome data are integrated. (Mohammed Fatih Rasul, 2022).
The advancement of genetic engineering instruments in cancer treatment, emphasizes the drawbacks of previous approaches such as ZFNs and TALENs. It presents CRISPR/Cas9 as a more flexible method that targets and recognizes sequences using RNA molecules. The Class 1 and Class 2 CRISPR/Cas9 systems are divided according to how they recognize and cleave viruses and plasmids. The CRISPR/Cas9 system is derived from prokaryotic defensive mechanisms against these pathogens. Class 2 systems provide potential targets for genome engineering, such as Type II, V, and VI. Nevertheless, certain targeted constraints exist, such as the identification of protospacer adjacent motifs (PAMs) and protospacer flanking sequences. The extensively researched CRISPR protein Cas9 operates as an RNA-guided endonuclease that needs trans-activating crRNA (tracrRNA) in order to recognize and cleave targets. Genome editing is made easier by the union of tracrRNA and crRNA into a single molecule called single guide RNA (sgRNA).
Cas9 is an enzyme that utilizes guide RNA, which is composed of tracrRNA and crRNA, to cut particular DNA regions. While tracrRNA helps Cas9 connect to the DNA, crRNA matches the target DNA sequence.
Figure 4
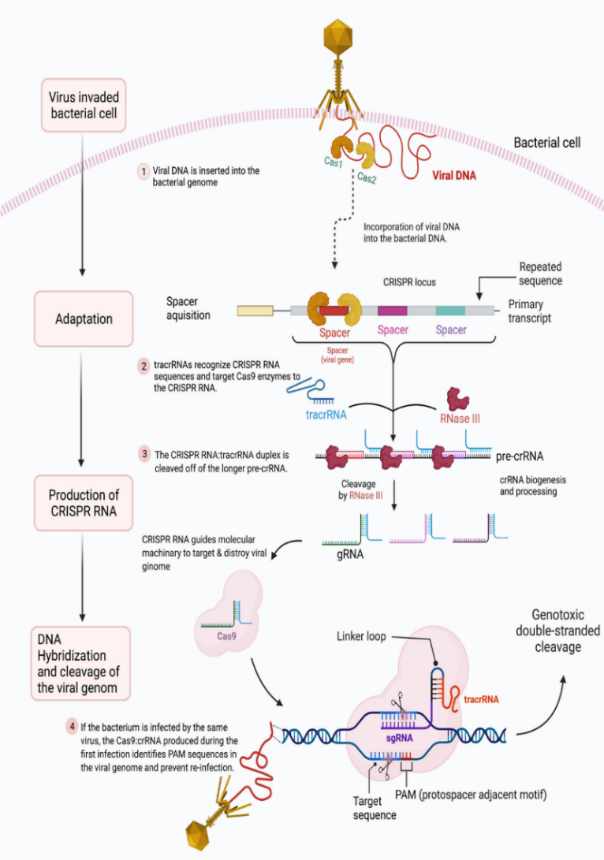
1. Adaptation: When phage DNA penetrates a bacterial cell, the Cas1-Cas2 proteins become active. As a result, phage DNA fragments are eliminated and integrated into the bacterial genome's CRISPR array (Mohammed Fatih Rasul, 2022).
2. Biogenesis: Pre-crRNA is produced when the CRISPR array is transcribed, and it is later transformed into mature crRNA.
3. Interference: Effectors of the cas protein bind to crRNAs. In the event that a phage re-invades and its sequence aligns with a crRNA, Cas effectors utilize an R-loop complex formation and Cas executor nuclease activity to eliminate the phage DNA.
Innovative Advances in CRISPR/Cas9 Gene-Editing Technology
Unaware of their importance, Japanese researchers found tandem repeats in the E. coli genome in 1987. Later, in 2002, these repeats were dubbed CRISPR. The involvement of CRISPR loci in adaptive immunity was discovered in 2005. In 2007, Barrangou's group demonstrated that viral gene sequences incorporated into the bacterium's genome could alter the bacterium's resistance to phages. Subsequent investigation by Brouns et al. in 2008 showed that Cas proteins might be directed to target DNA for protection by non-coding RNA from CRISPR. The 2008 discovery by Deltcheva et al. of tracrRNA's function in pre-crRNA processing added to our knowledge of crRNA formation. Research published in 2012 showed how mature tracrRNA and crRNA directed Cas9 to cleave DNA. When the Cong and Mali teams enabled genome editing with CRISPR/Cas9 in cell cultures in 2012, the field experienced significant advancements. By 2020, a plethora of CRISPR/Cas9-based gene editing instruments have been developed as a result.
Figure 5
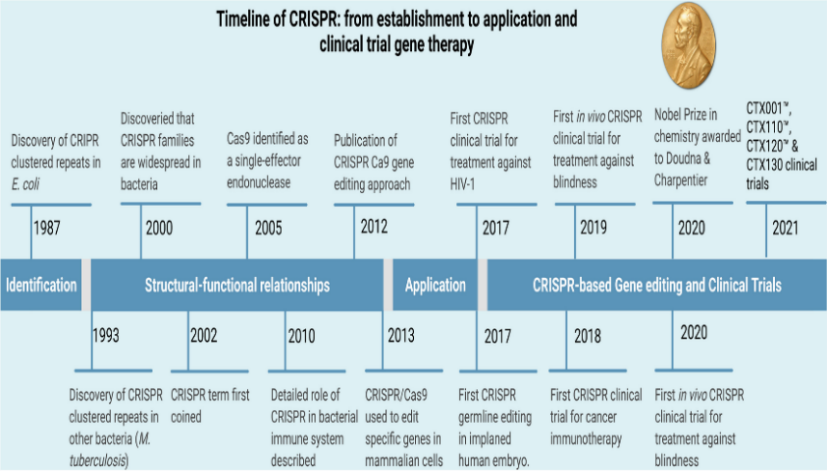
A timeline that highlights the key discoveries, applications, structural-functional linkages in CRISPR development, CRISPR-based gene editing, and clinical trials
Discussion about CRISPR/Cas9-Based Genome Technology
About 50% of bacteria have a defense mechanism called the CRISPR/Cas system that aids in the bacterial defense against viral infections. It targets and obstructs particular viral sequences with the use of small guide RNAs. The system is made up of a genomic region known as CRISPR that has distinct spacer sequences derived from viruses and repeating elements. Cas genes, which produce the Cas proteins, are normally located close to an AT-rich area at the locus's start. Based on the composition and roles of the Cas proteins, the two primary classes of CRISPR/Cas systems are class I and class II, which are further split into six types (types I–VI). Nucleic acid cleavage is carried out by multiprotein complexes in class I systems.
Figure 6

Since only the Cas9 protein is utilized in Type II CRISPR systems, they are more efficient than multiprotein strategies. This approach, which is frequently employed in research, uses Cas9 to recognize and cut the DNA target sequence. The epigenome can be targeted by deactivated Cas9 without causing sequence disruptions. Two pieces make up the guide RNA: a spacer detects and attaches to the main site, while the other component attaches to the Cas protein. Near the cut site, a PAM sequence aids in identifying the target DNA for the introduction of a mutation. (Mohammed Fatih Rasul, 2022).
Clinical Trials and the Use of CRISPR/Cas9 Gene Editing for Anticancer Purposes
Since Type II CRISPR systems solely employ the Cas9 protein, they are more efficient than multiprotein strategies. In this technique, which is frequently employed in research, the DNA target sequence is identified and cleaved by Cas9. Sequences cannot be broken by deactivated Cas9, which can target the epigenome. Two pieces make up the guide RNA; one binds to the Cas protein, while the other, a spacer, finds and attaches to the target site. A PAM sequence that is close to the cut site aids in locating the target DNA so that a mutation can be introduced. Clinical trials (Table 2) examining the effectiveness of CRISPR-based cancer therapies in addressing cancer-causing genes have been made possible by encouraging pre-clinical findings.
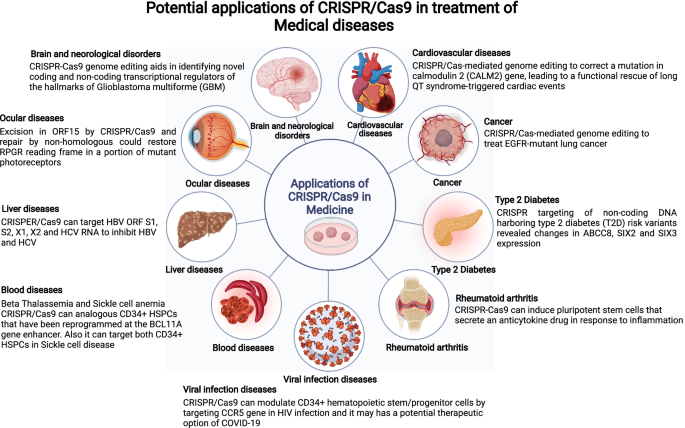
Recent clinical experiments have shown how CRISPR/Cas9's potential to treat a wide range of disorders is growing. A wide range of diseases, including viral infections, neurological disorders, cancer, ophthalmic ailments, blood disorders, cardiovascular diseases, and other complex genetic conditions have been modeled using this ground-breaking method.
Targeting the PD-1 protein in cancer treatment shows promise in enhancing the immune system's response. The PD-1 inhibitor pembrolizumab has been successful in the therapy of non-small cell lung cancer (NSCLC). Furthermore, the CRISPR/Cas9 technique is being investigated to stop the expression of PD-1 in cancer cells, indicating promise for customized immunotherapy. (Mohammed Fatih Rasul, 2022).
The paragraph discusses the ongoing clinical trials and advancements in cancer treatment using CRISPR gene editing technology. PD-1 knockout-engineered immune cells have been directed against cancers of metastatic NSCLC, PD-1 deletion in T cells against cancers of renal, bladder, prostate, esophagus, and others. The generation of chimeric antigen receptor T cells—CAR-T cells that have been designed against antigens CD19 and CD20/CD22 in leukemia, is another example of how the CRISPR gene editing technique has been used. In addition, it outlines the therapy of cervical cancer with the help of CRISPR/Cas13a, in which HPV 16/18 E6/E7 mRNAs are brought to a standstill. Finally, it enlists some of the clinical trials that are underway; some of these include one for Neurofibromatosis type 1 (NF1) by using induced pluripotent stem (Mohammed Fatih Rasul, 2022).
Figure 7
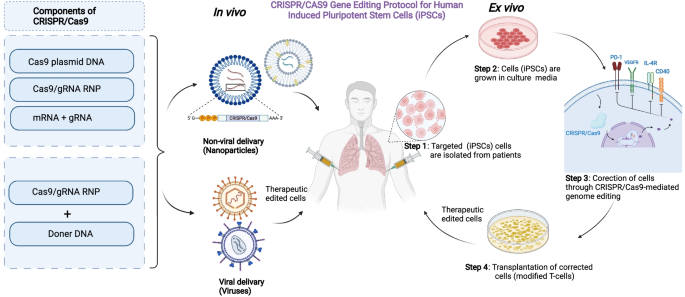
It has recently made it possible to use CRISPR/Cas9 gene editing techniques to manipulate genes in human-induced pluripotent stem cells both in vitro and in vivo, enabling the creation of isogenic control iPSC lines. These studies have helped in understanding the genetic pathways pertinent to diseases and cellular functions.
The NF1 genetic states were used in the identification of drug targets for generating cell lines by use of the CRISPR/Cas9 system. Though this system is quite encouraging in this respect, further research has to be done to ascertain its safety and efficacy in cancer treatment. Furthermore, owing to the high sensitivity and speed in cancer therapy, such drug-resistance mutations as FLT3-F691L can be rapidly identified by CRISPR/Cas9.
Challenges of CRISPR/Cas9
The CRISPR/Cas9 gene-editing technique has a lot of potential, but it still has a lot of drawbacks and hazards that will affect how it is used in clinical trials. Concerns include immunogenicity, off-target effects, polymorphism, challenges with delivery, and moral dilemmas. Different approaches have been developed to deal with these problems, as Figure 5 illustrates.
Figure 8
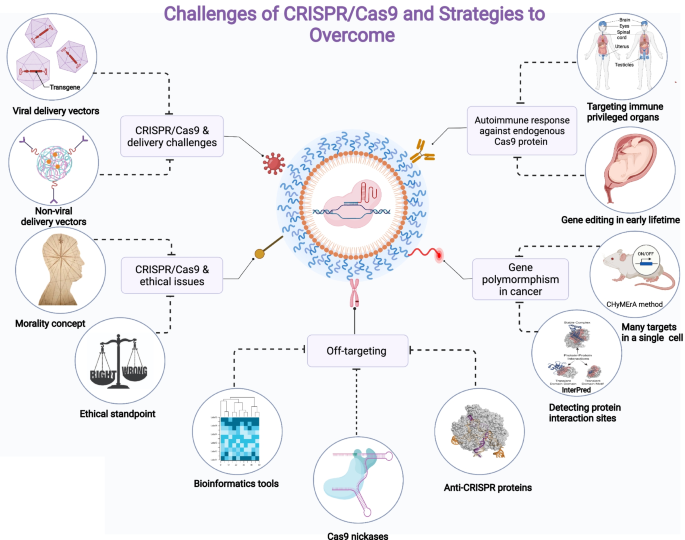
Clinical studies and human applications of CRISPR/Cas9 provide a number of challenges. Immunogenicity, off-target effects, genetic diversity, delivery strategies, and ethical issues are among the main obstacles. To overcome these challenges, plans for reducing immune reactions, improving precision targeting, accommodating genetic differences, streamlining delivery methods, and addressing ethical issues must be developed. (Mohammed Fatih Rasul, 2022).
The reaction of the Immune System to the Natural Cas9 Protein
The Cas9 protein, a key element of CRISPR systems, attaches to and cuts specific regions of DNA. Originating from bacteria such as Staphylococcus aureus and Streptococcus pyogenes, it causes immunological reactions in people because of previous infections. Because of the immune system's detection of it, gene editing efficacy may be hampered by its quick breakdown after injection.
Strategies to Overcome Immunogenicity
Numerous approaches have been put out to get over the restrictions brought on by immunogenicity against Cas9. Here, we're summarizing the primary options available; (i) using the CRISPR/Cas system to change genes at a young age; Targeting organs with immune privileges (ii) (Fig. 6).
Figure 9
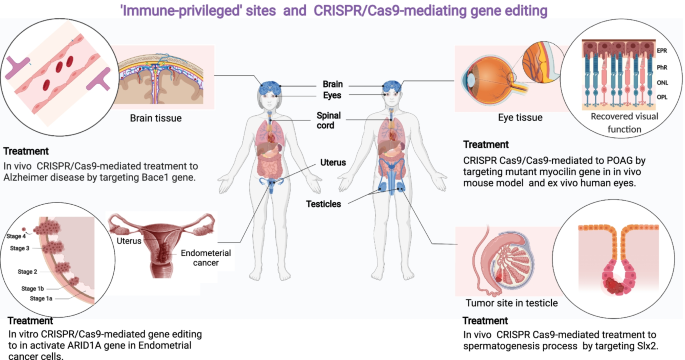
Scientists are investigating two approaches to address the problem of the immune response to Cas9: targeting immune-privileged organs and employing CRISPR/Cas9 gene editing at an early age. These strategies seek to get beyond the restrictions brought forth by the way the immune system reacts to Cas9.
Gene Editing in Early Lifetime
Prenatal diagnosis of pediatric illnesses is possible, and the CRISPR/Cas system has shown promise in the treatment or prevention of some genetic illnesses, including sickle cell anemia, thalassemia, mucopolysaccharidosis type IVA, and cystic fibrosis. Furthermore, CRISPR/Cas9 has been successful in addressing molecular pathways linked to prevalent children's malignancies like lymphoma and neuroblastoma. It is possible to treat these disorders using CRISPR/Cas before the infant receives a vaccination against the Cas protein.
Targeting Immune-Privileged Organs
Targeting immune-privileged tissues including the brain, testicles, and eyes with CRISPR/Cas9 gene editing has shown promise in recent years for correcting genetic abnormalities. For example, it's been effectively applied to decrease the expression of defective genes in eye cells, which may help treat disorders such as Leber congenital amaurosis. Studies have indicated that off-target rates can reach 16%, which raises serious concerns about off-target effects. Gene alterations that are not planned can result from off-target effects. Although bioinformatics techniques are now able to predict and eliminate off-target effects, additional developments are required to increase their efficacy in the development of novel medicines. (Mohammed Fatih Rasul, 2022).
Techniques to Prevent Off-targeting
Previous research has effectively employed three primary approaches: (I) integrating anti-CRISPR proteins; (II) employing Cas9 nickases to efficiently create precise gRNA and forecast off-target effects; and (III) using bioinformatics techniques.
Technologies of Bioinformatics
Bioinformatics tools are critical for studying the CRISPR-Cas system. Francisco Mojica was aided in determining that it was present in Archaea. These approaches facilitate the accurate localization of editing sites, the efficient creation of gRNAs, and the mitigation of off-target effects. Studies reveal that gRNA length influences off-targeting, with shorter lengths significantly reducing the probability. To retain efficiency, gRNAs can be shortened to less than 20 nucleotides, which can reduce off-targeting by up to 5000 times. Since the majority of mismatches occur in the final three nucleotides opposite the PAM region, removing mismatches and keeping the gRNA at around 17 nucleotides can reduce off-targeting. However, gRNAs with fewer than 15 base pairs may become less selective and unable to bind the intended target within the nucleus.
Cas9 Nickases
A helpful method for mitigating off-target effects in gene editing is utilizing CRISPR nickase to modify a single nuclease domain within a single DNA strand. Cas9 nickase uses double adjacent gRNAs and breaks just one strand of DNA, in contrast to ordinary Cas9. This method, shown in Figure 7, lowers the chance of off-target damage by lessening DNA damage in the target area. Research indicates that matched nicking can reduce off-targeting in cell lines by 50–1500 times and allow for gene deletion in mouse zygotes without sacrificing cleavage efficiency.
DNA cutting requires the Cas9 endonuclease protein's amino acids H840 and D10. D10 in the HNH domain cuts the gRNA-targeted strand, while H840 in the RuvC domain cuts the non-targeted DNA strand. Cas9 only cuts the gRNA-complementary strand when employing single nickases; however, when using paired nickases, both strands can be nicked at the same time. A 5' overhang, gRNA spacing, and the respective target site placements of each gRNA are all factors to be taken into account when designing gRNAs for paired nickases.
Figure 10

Anti-Crispr Proteins
After Cas9 has targeted a site, it can be deactivated to reduce its efficacy off-target. To date, more than 50 different types of anti-CRISPR proteins (Acr) have been found; these proteins deactivate Cas9. Because Acr proteins are tiny and varied, it is challenging to identify them. To deactivate CRISPR/Cas9, they employ a number of strategies, including attaching to both Cas9 and sgRNA. Acr efficiency is influenced by the binding of DNA and concentration. Conversely, Aca proteins stop Acr transcription. Drug resistance may be aided by the use of Acr proteins rather than antibiotics.
Screening Before the Treatment
Genes such as TP53 and KRAS that already have mutations may be more susceptible to new alterations during CRISPR Cas cancer therapy [166]. The two main strategies to address this issue are patient monitoring following injection and screening prior to employing the CRISPR Cas system.
Cancer Polymorphism
By restoring the function of tumor suppressor genes that
have undergone mutations, CRISPR Cas9 presents a possible treatment path for cancer by impeding the growth of new tumors. However, repairing single nucleotide alterations is difficult because of the complexity of cancer genetics. Although accurate, knocking in altered nucleotides is time-consuming and requires many guide RNAs. As an alternative, oncogenes like KRAS and ATM can be interfered with by CRISPR Cas9 by removing their inactivation sequences. Deactivating certain genes, such as CXCR7 in TNBC cells, may be able to prevent the activation of oncogenes in certain situations. To effectively disrupt important biochemical pathways in cancer progression, accurate gRNA design is necessary for effective CRISPR targeting of oncogenes.
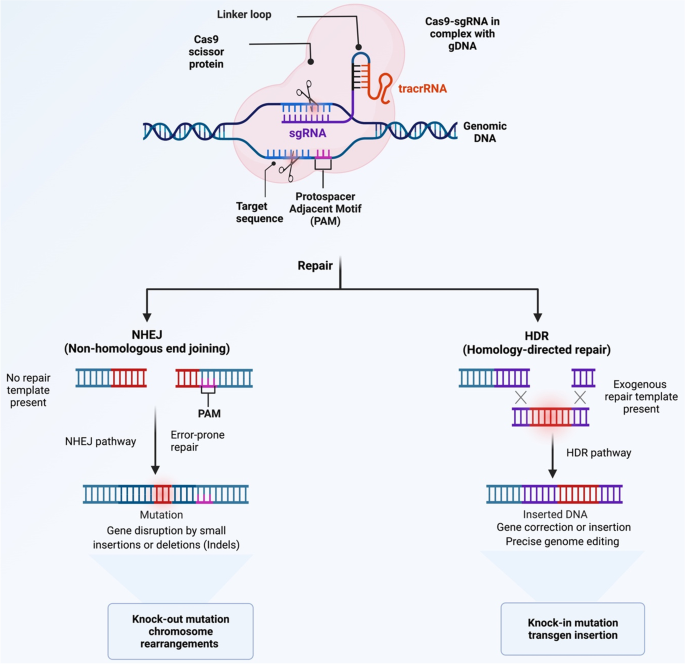
When CRISPR/Cas9 damages DNA, it is often repaired using non-homologous end joining (NHEJ) or homology-directed repair (HDR). While NHEJ results in random mutations (indels) that might alter gene sequences or reorganize the genome, HDR uses external repair templates to fix DNA. The results of DNA repair can be influenced by pushing cells to prefer one repair method over another by employing certain chemicals. (Mohammed Fatih Rasul, 2022).
Figure 11
Strategies
The two main approaches that were discussed to address polymorphism issues were protein interaction site analysis and prediction using bioinformatics techniques, such as CHyMErA. Here's a condensed overview: We discussed two approaches to polymorphism problems: the use of bioinformatics to research protein interactions and forecast outcomes, and the use of CHyMErA. (Mohammed Fatih Rasul, 2022).
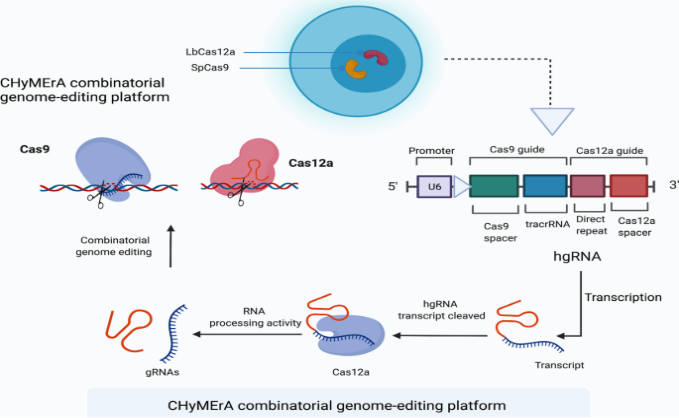
CHyMErA Experimentation
Different CRISPR strategies are used by researchers to efficiently edit numerous targets within a single mammalian cell, such as CHyMErA (Cas hybrid for multiplexed editing and screening applications). Both the Cas9 and Cas12a nucleases are utilized by the CRISPR/Cas9 system that CHyMErA employs. Exons can be deleted with this technique, which makes it possible for gene sequences to be deleted at high rates. A possible approach to treating cancer polymorphism is to use CHyMErA to target numerous locations.
CHyMErA is a platform for editing DNA that employs multiple components. It involves cell lines with the enzymes SpCas9 and LbCas12a as well as hgRNA expression cassettes. This method uses the same U6 promoter seen in hgRNAs to express Cas12a gRNAs alongside Cas9. Functional Cas9 and Cas12 gRNAs are released by Cas12a upon recognition and cutting of a specific sequence. These can then be coupled to achieve accurate genome editing.
Figure 12
Detecting Protein Interaction Site
Detecting Protein Interaction Site
Bioinformatics tools such as InterPred help find protein interaction sites during CRISPR Cas9 gene editing, which is important for generating gRNAs. Numerous databases aid in the prediction of CRISPR knockout results. Although accuracy is crucial when modifying certain nucleotides through knock-ins, methods such as ChyMErA manage multi-target alterations by utilizing Cas9 and Cas12a. On the other hand, cancer therapy is more feasible when oncogenes' active site mutations are eliminated because this effectively suppresses their function. (Mohammed Fatih Rasul, 2022).
The Delivery Challenges
Choosing an appropriate and accurate delivery method for the CRISPR system in vivo poses obstacles. There are three types of delivery systems: extracellular vesicle-based, physical, and viral. Each has advantages and disadvantages. Although lentivirus and adenovirus are common viral vectors, they have limitations in terms of package size and immune response. Adeno-associated viruses (AAVs), for example, need several vectors in order to efficiently transport all of the CRISPR components due to their small package size. Continuous exposure raises the chance of mutations and off-target consequences even in the case of a modest immune response.
Non-Viral Delivery Vectors
Non-viral delivery techniques, like lipid and inorganic nanoparticles, have benefits for gene therapy, including decreased immunogenicity, decreased exposure to enzymes, and more accurate targeting. These methods enable less risky, frequent administration. Non-viral vectors can transport greater payloads without integrating into the genome, in contrast to viral vectors. Extracellular vesicle-based systems are safe, and affordable, and have shown promise in both in vitro and in vivo applications. Nevertheless, difficulties still exist in delivering CRISPR systems effectively, particularly in cancer treatment, where perfect editing efficiency is still a barrier. For example, using hydrodynamic injection to target liver cells results in a delivery efficiency of only 1 in 250. (Mohammed Fatih Rasul, 2022).
Strategies
Cas9 protein is divided into two AAV vectors, and the packaging problem in genetic editing can be effectively addressed. Off-targeting hazards are increased by large vectors; this risk is mitigated by utilizing smaller Cas9 segments spaced between AAV vectors. Utilizing ribonucleoprotein complexes such as CRISPR-Cpf1-RNP, which reduce off-targeting, is a further technique. According to studies, Cas9-RNP techniques are highly effective in cell cultures and spontaneously break down in 24 to 48 hours, reducing the possibility of long-term alterations and off-targeting that come with viral vectors (Mohammed Fatih Rasul, 2022).
CRISPER Technology and its Ethical Problems
Given that the CRISPR/Cas9 genome editing technique has the ability to alter human DNA, it has generated a number of ethical discussions in recent years. Its promising applications entail unpredictable consequences and call for careful consideration of its ethical and social impacts. As such, insights from diverse perspectives—from the public to religious scholars—should first be sought in sustaining a comprehensive scrutiny of the implications of the technology.
Current Moral Position
The utilization of the CRISPR/Cas9 technology is fast growing in molecular biology, having gene manipulation for research and practical applications in crop improvement and breeding of stronger animals. However, it will mean a possible altering of human genomes to prevent diseases or to enhance human beings, which raises concerns relating to ethical issues since modification of germ cells is inheritable. These concerns have in turn raised calls for the reaching of a global consensus on the ethical and safety implications of its use prior to starting human germline editing.
Aspects in Morality
Decisions in biomedical ethics, including those related to technologies like CRISPR, are made based on a balance of risks against potential benefits. Some of the problematic issues in regard to editing with CRISPR are efficiency and precision. It is still not clear what sort of effect such changes will have on species in the long term, or if such edited genes will be passed on to future generations. This makes it difficult to project what the outcome will be, and hence view risks, through limitations technically and biologically. Further, the way changes to genes impact traits is not fully understood; therefore, making a precise moral decision on this is even more difficult.
Summary of Strategies
In conclusion, patient safety must always come first when using CRISPR/Cas9 technology, as there are substantial potential advantages for health and wellness. It can potentially prevent the inheritance of genetic diseases, easing parental anxiety. Recent advancements, such as modifications to the Cas9 enzyme, aim to reduce off-target mutations, enhancing safety. Despite ongoing ethical and safety considerations, CRISPR/Cas9 holds promise for improving health and quality of life, contingent on responsible application.
Conclusion and Future Perspectives
Recent years have seen a transition from research into clinical trials for therapeutic genome editing, with an emphasis on editing hematopoietic stem cells (HSC), T cells ex vivo, and liver cells in vivo. Still, there are a lot of obstacles to overcome, with delivery being one of the biggest. Better delivery approaches are required since many tissues have low editing technique efficacies. International regulations should control gene editing experiments in healthcare systems to prevent potential harm (Mohammed Fatih Rasul, 2022).
Directions in the Future and its Current Status
The repurposing of CRISPR–Cas systems for eukaryotic cells has set off a revolution in genome engineering. RNA targeting techniques based on Cas13a are among the innovative technologies that have been produced by research into CRISPR systems from prokaryotic organisms, despite the widespread use of type II CRISPR-Cas systems. Combining dCas9 with different effectors might broaden the scope of targeted epigenetic modification. (Edward A. Stadtmauer, 2020).
Together with advances in next-generation sequencing, the more efficient creation of gRNA libraries for large-scale Cas9 targeting has created genome-wide genetic and epigenetic platforms. more accessible, greatly expanding our knowledge of biological mechanisms and making it easier to identify novel therapeutic targets. Furthermore, complex manipulation of biological processes is made possible by the multiplexed targeting capabilities of CRISPR Cas systems. (Adrian Pickar-Oliver, 2019).
With the advent of CRISPR-Cas-based medications in clinical trials, there is tremendous promise for improving cell treatments and curing genetic disorders. Preclinical results are promising, but throughout clinical trials, caution about safety and efficacy is crucial. Reducing the likelihood of off-target genomic modifications requires improving techniques for identifying uncommon mutations and evaluating their possible consequences, which is essential for the advancement of CRISPR-based therapies. (Edward A. Stadtmauer, 2020).
Cite this article
-
APA : Masood, A. J., Ahmed, E., & Iftikhar, R. (2024). Applications and Challenges of CRISPER/Cas-9 System in Cancer Therapy. Global Pharmaceutical Sciences Review, IX(I), 10-28. https://doi.org/10.31703/gpsr.2024(IX-I).02
-
CHICAGO : Masood, Ahmad Jalal, Ejaz Ahmed, and Rimsha Iftikhar. 2024. "Applications and Challenges of CRISPER/Cas-9 System in Cancer Therapy." Global Pharmaceutical Sciences Review, IX (I): 10-28 doi: 10.31703/gpsr.2024(IX-I).02
-
HARVARD : MASOOD, A. J., AHMED, E. & IFTIKHAR, R. 2024. Applications and Challenges of CRISPER/Cas-9 System in Cancer Therapy. Global Pharmaceutical Sciences Review, IX, 10-28.
-
MHRA : Masood, Ahmad Jalal, Ejaz Ahmed, and Rimsha Iftikhar. 2024. "Applications and Challenges of CRISPER/Cas-9 System in Cancer Therapy." Global Pharmaceutical Sciences Review, IX: 10-28
-
MLA : Masood, Ahmad Jalal, Ejaz Ahmed, and Rimsha Iftikhar. "Applications and Challenges of CRISPER/Cas-9 System in Cancer Therapy." Global Pharmaceutical Sciences Review, IX.I (2024): 10-28 Print.
-
OXFORD : Masood, Ahmad Jalal, Ahmed, Ejaz, and Iftikhar, Rimsha (2024), "Applications and Challenges of CRISPER/Cas-9 System in Cancer Therapy", Global Pharmaceutical Sciences Review, IX (I), 10-28
-
TURABIAN : Masood, Ahmad Jalal, Ejaz Ahmed, and Rimsha Iftikhar. "Applications and Challenges of CRISPER/Cas-9 System in Cancer Therapy." Global Pharmaceutical Sciences Review IX, no. I (2024): 10-28. https://doi.org/10.31703/gpsr.2024(IX-I).02
